The number of HLA alleles has been growing exponentially over the last couple of years (it just reached 30 000 in April 2021, IMGT 3.44, figure 1) and it looks like the curve is not stopping any time soon, which does not make the lives of scientists in the transplant field easy. Several approaches have been established to simplify the process of finding a best matched transplant and to make the research more “user friendly“. One of them is following the task that Cinderella had before she was allowed to go to the ball and meet her Prince – the grain sorting – classifying HLA alleles into big groups called Supertypes.
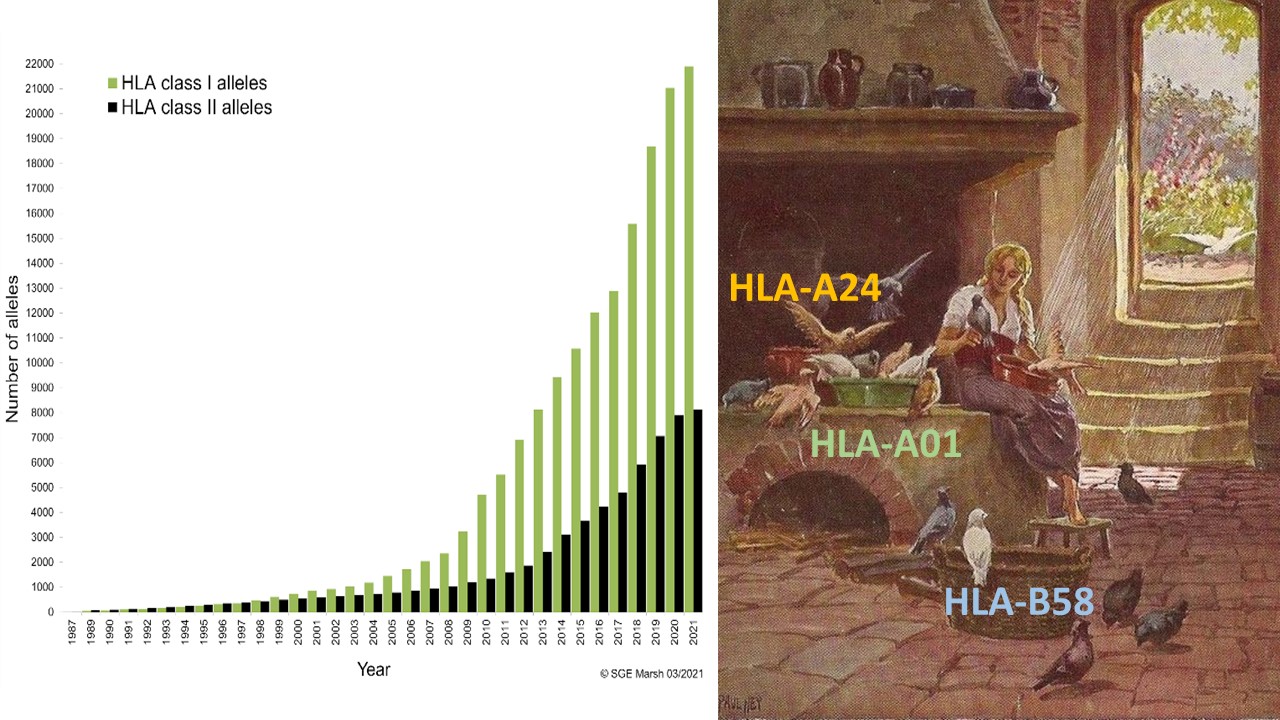
Figure 1. Number of alleles named by year from 1987 to 2021 (1).
The concept of supertypes was firstly introduced by Alessandro Sette’s group in 1995. The definition of an HLA supertype is that HLA molecules with similar peptide binding features are grouped into one supertype, meaning that if a peptide is able to bind to one allele within a supertype, it can also bind to all other alleles in this supertype. This group focused on HLA-A and -B loci and defined 9 supertypes (HLA-A1, -A2, -A3, -A24, -B7, -B27, -B44, -B58, -B62), which were reported to cover most of the HLA-A and -B polymorphisms. The classification was based on clustering the structural motifs derived from experimentally-determined binding data (2).
In 2008, the group revised and updated the original classification based on the IMGT database (at that time IMGT 2.9) and used an approach based on compilation of published motifs, binding data and analysis of shared repertoires of binding peptides, in combination with clustering based on the primary sequence of the B and F peptide-binding pockets of Class I molecules (Figure 2) (3).
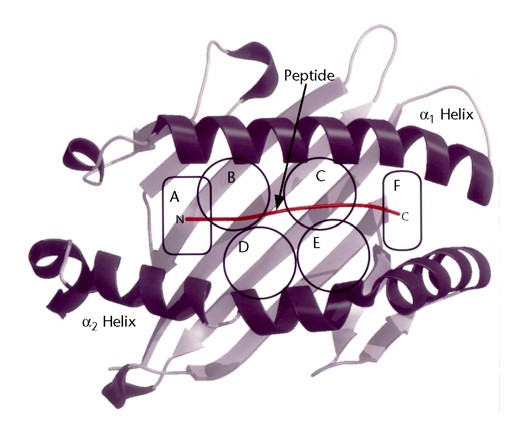
Figure 2. Six pockets defined in the peptide-binding cleft of MHC class I molecules (4).
In the end, 80% of the 945 different HLA-A and -B alleles could be assigned to one of the original 9 supertypes. However, they found that some alleles have specificities spanning over two different supertypes and proposed to increase the number to 12, adding A01 A03, A01 A24 and B08 category (summary of HLA supertype description, table 1) (3).
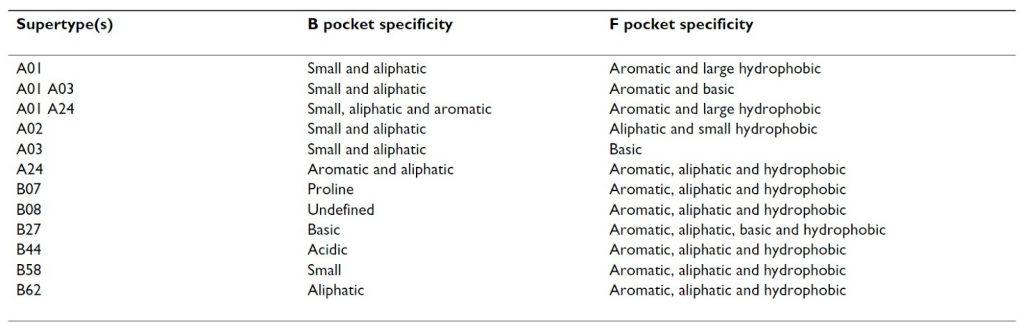
Table 1. HLA supertype specificity descriptions (3).
Multiple groups have been focusing mostly on the Class I supertype approach, trying to further define and improve the classification. In the end, the updated HLA Class I classification described by Alessandro Sette’s group was in agreement with those defined by the approaches from the other groups (e.g. Ole Lund’s group) and is now widely accepted (5,6).
In contrast to HLA Class I supertypes, the number of studies about Class II supertypes has been much lower. One important reason is that peptide binding data for Class II molecules is less available than those for Class I molecules due to the complexity of Class II structure. Nevertheless, the studies have suggested the similarity that many DR molecules and many DP molecules can be grouped into supertypes (7,8).
One of the examples of implementing the supertype approach is a study performed by Chowell et al. who investigated the clinical relevance of individual HLA Class I alleles after immune checkpoint blockade therapy. The group examined the effects of Class I supertypes on overall survival and identified two B supertypes, associated with survival outcome in patients with advanced melanoma in two independent cohorts. Patients with B44 superfamily alleles had significantly better survival and, in contrast, patients carrying B62 alleles had significantly reduced survival. Besides the individual effect of supertypes, they also focused on the impact of zygosity confirming that maximal heterozygosity at HLA Class I loci improved overall survival. The conclusion was that their results have important implications for predicting treatment response suggesting these factors could be considered in the design of future clinical trials even if further experimental work is necessary to test some of their additional hypotheses (9).
Following this research, Camacho-Bydume et al. investigated the impact of HLA heterozygosity, supertypes and surface expression on outcomes in adult and pediatric patients with myeloid and lymphoid malignancies undergoing unrelated hematopoietic cell transplantation (HCT). Unlike in the above-mentioned paper, HLA Class I heterozygosity and also HLA expression were not associated with overall survival or other post-transplant outcomes such as relapse, transplant-related mortality (TRM), disease-free survival (DFS) and acute graft-versus-host disease. Nevertheless, they confirmed that B62 supertype was associated with decreased TRM in the entire patient cohort and that B27 supertype was associated with worse DFS in patients with AML which was a unique finding in their study and may have prognostic utility. Their conclusion was that the survival benefit of HLA heterozygosity seen in solid tumor patients receiving immune checkpoint inhibition does not extend to patients undergoing allogeneic HCT. However, certain HLA supertypes are associated with TRM and DFS, suggesting that similarities in peptide presentation between supertype members play a role in these outcomes. Beyond implications for prognosis following HCT, these findings support the further investigation of these HLA supertypes and the specific immune peptides important for transplant outcomes (10).
There have been plenty of publications involving stratification of HLA alleles based on the just described approach where HLA supertypes have been associated with multiple disease associations and various outcomes. Nevertheless, over the time there have also been a number of other Cinderella-like paths which aim to classify HLA alleles into clusters or supertypes. Despite the differences in various classification schemes, the concept of HLA supertypes has been involved in multiple disease studies. It has been effectively used to characterize and identify T cell epitopes and also utilized as a component in several approaches and algorithms designed for predicting peptide binding capacity.
Having its limitations, the HLA supertype approach has been used to continuously investigate the HLA fairytale and, unlike for Cinderella, there are more balls waiting for the HLA community with one of them happening next year in Amsterdam 😉
References:
- Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SGE The IPD and IMGT/HLA database: allele variant databases Nucleic Acids Research (2015) 43:D423-431, https://www.ebi.ac.uk/ipd/imgt/hla/
- Sidney J, Grey HM, Kubo RT, Sette A: Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol Today 1996, 17:261-6
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1.
- Achour, A. (2001). Major Histocompatibility Complex: Interaction with Peptides. In eLS, (Ed.). https://doi.org/10.1038/npg.els.0000922
- Lund O, Nielsen M, Kesmir C et al (2004) Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 55:797–810
- Tong JC, Tan TW, Ranganathan S (2007) In silico grouping of peptide/HLA class I complexes using structural interaction characteristics. Bioinformatics 23:177–183
- Greenbaum J, Sidney J, Chung J et al (2011) Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 63:325–335
- Saha I, Mazzocco G, Plewczynski D (2013) Consensus classification of human leukocyte antigen class II proteins. Immunogenetics 65:97–105
- Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582–587.
- Camacho-Bydume C at al. Specific Class I HLA Supertypes but Not HLA Zygosity or Expression Are Associated with Outcomes following HLA-Matched Allogeneic Hematopoietic Cell Transplant: HLA Supertypes Impact Allogeneic HCT Outcomes. Transplant Cell Ther. 2021 Feb;27(2):142.e1-142.e11. doi: 10.1016/j.bbmt.2020.10.010






