
-
ONE report in <3 hours!
-
ONE test delivers both
absolute and relative quantities -
Scalable from ONE to
many samples -
The same ONE test
every time -
ONE test provides more
information than
NGS for less
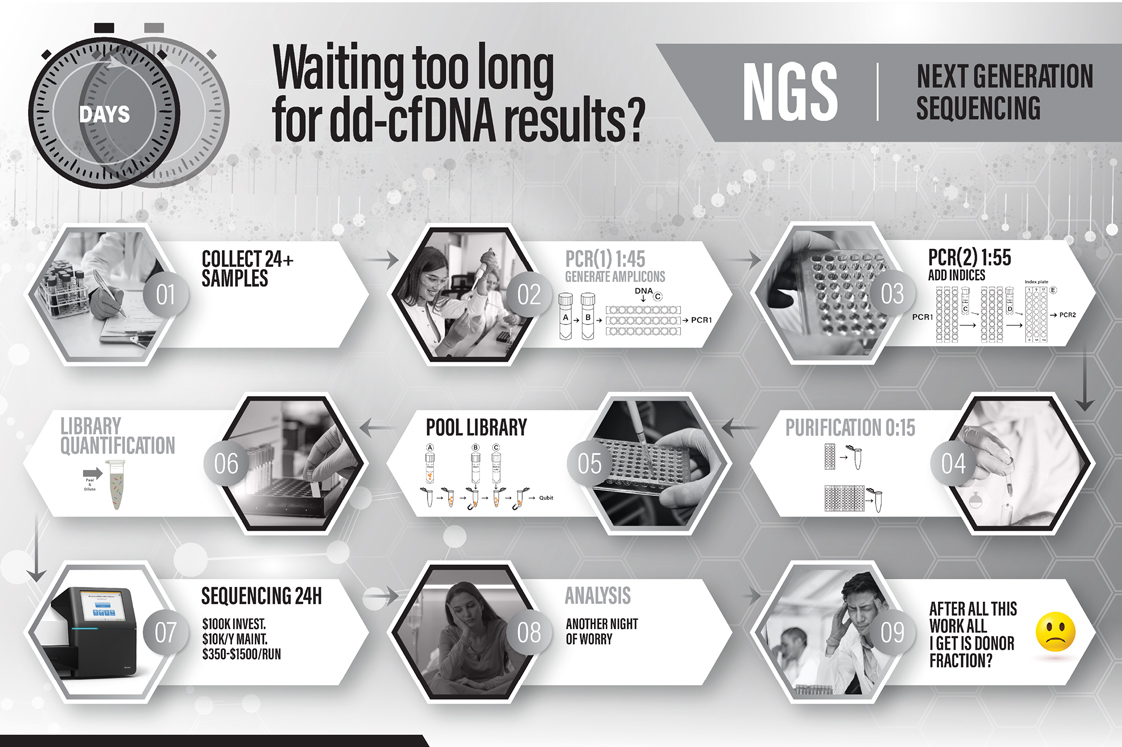
How does HoloGRAFT ONE make frequent dd-cfDNA monitoring feasible and cost-effective?
Frequent dd-cfDNA monitoring in transplantation may allow early detection of allograft injury, which is expected to improve graft survival rates, and reduce the need for unnecessary invasive procedures. This unique biomarker has the promise of improving the monitoring of response to treatment or changing therapy.
The simplicity of our unique HoloGRAFT ONE Assay and Software, which utilizes digital PCR, reduces both time and expense traditionally associated with dd-cfDNA monitoring. The simplicity and affordability of HoloGRAFT ONE workflow makes it suitable for frequent monitoring of dd-cfDNA.

- HoloGRAFT ONE eliminates the need for prior genotyping of the donor and recipient.
- Digital PCR is economical even for single-sample runs.
- HoloGRAFT ONE simultaneously measures all markers, and the test provides a comprehensive evaluation of dd-cfDNA levels.
- Direct absolute quantification of dd-cfDNA by digital PCR eliminates the need for complex library preparation, pre-amplification and expensive NGS instrumentation.
- HoloGRAFT ONE measures dd-cfDNA level solely from the patient’s sample.
Background Information
HoloGRAFT ONE uses copy number variant markers, long stretches of DNA inherited in zero, one or two-copy forms. The donor-informative markers, as well as their detected amplicons, are missing in the patient. The detected amplicons are subsets of the variants, in contrast to SNP-based alternatives, where the variants are subsets of the detected amplicons. Therefore, HoloGRAFT ONE is the only dd-cfDNA monitoring method with a negative background.
• The accumulated evidence supporting the use of dd-cfDNA argues that its use should no longer be considered investigational. The use of donor-derived cell-free DNA (dd-cfDNA) is evidence-based and validated, and a prime example of much needed innovation in transplant organ surveillance. It has demonstrated utility in the early detection of allograft injury and assistance with clinical decision-making regarding allograft biopsy and treatment initiation. (ASTS Position Statement)1
• dd-cfDNA is a better discriminator of acute kidney rejection than serum creatinine. (Bloom et al, 2017) 2
• Elevated dd-cfDNA levels are associated with increased likelihood of de novo DSA development in kidney recipients.(Jordan et al. 2018)3
• Elevated dd-cfDNA levels were found to predict longitudinal eGFR decline, while normal levels did not have evidence of eGFR decline during the period of study follow-up. (Stites et al. 2020)4
HoloGRAFT ONE is the only method of direct
absolute quantification of dd-cfDNA.
Direct and absolute quantification of dd-cfDNA involves the direct detection and counting of the dd-cfDNA molecules present in the sample without pre-amplification. Using HoloGRAFT ONE the precise number of dd-cfDNA molecules present in the sample can be determined, enabling absolute quantification.
Frequent dd-cfDNA monitoring in transplantation may allow early detection of allograft injury, which is expected to improve graft survival rates, and reduce the need for unnecessary invasive procedures. This unique biomarker has the promise of improving the monitoring of response to treatment or changing therapy.
The simplicity of our unique HoloGRAFT ONE Assay and Software, which utilizes digital PCR, reduces both time and expense traditionally associated with dd-cfDNA monitoring. The simplicity and affordability of HoloGRAFT ONE workflow makes it suitable for frequent monitoring of dd-cfDNA.
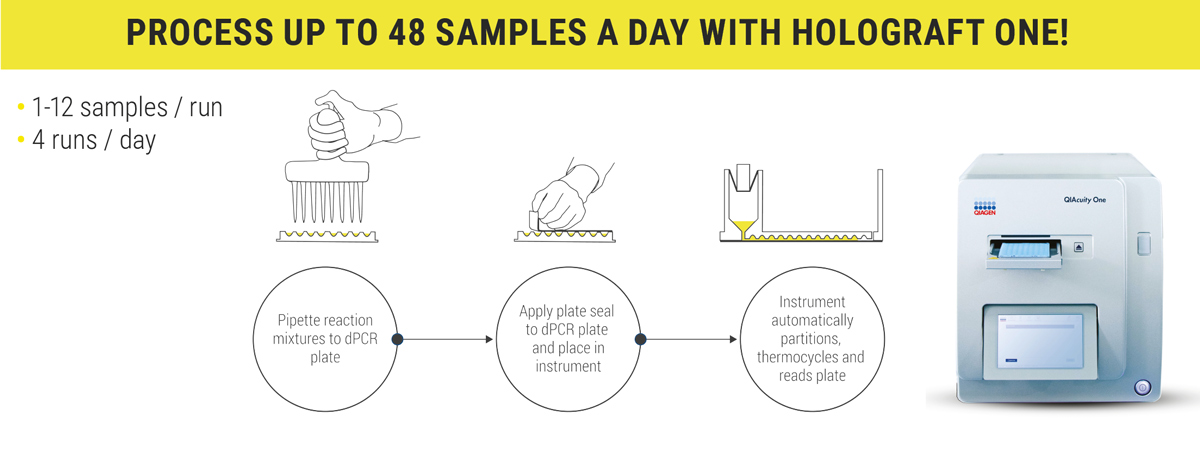
HoloGRAFT ONE is the fastest and
scalable dd-cfDNA monitoring method.
HoloGRAFT ONE is dPCR assay (digital polymerase chain reaction assay) based upon self-quenched, hydrolysis probe chemistry. Each assay is pre-mixed with an RNaseP control gene assay, in order to quantify both the amount of donor cfDNA and the total cfDNA in a single reaction.
Compared to traditional PCR methods multiple samples can be simultaneously analyzed in a single dPCR run, that saves time by minimizing the setup and processing steps.
1 ASTS Statement on donor-derived cell-free DNA (dd-cf-DNA) Approved by the ASTS Executive Committee on March 6, 2023 https://asts.org/docs/default-source/position-statements/dd-cfdna-position-statement.pdf
2 Bloom RD, Bromberg JS, Poggio ED, et al. Cell-free DNA and active rejection in kidney allografts. JASN. 2017;28:2221-2232.
3 Jordan SC, Bunnapradist S, Bromberg JS, et al. Donor-derived cell-free DNA identifies antibodymediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct. 2018. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6133406/.
4 Stites E, Kumar D, Olaitan O, et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant. 2020;20:2491-2498 PMID 32056331.

Take The Leap To Affordable, Frequent dd-cfDNA Monitoring
Please contact holograft@omixon.com and we will provide you with all the necessary information.

