The three “bigs” (in no particular order), with some links, basic information collected from the linked websites** and my (subjective) one sentence summary:
Illumina
- Technology videos:
-
HiSeq 2000 (shows the workflow)
-
Reads:
-
Read length: 25, 36, 50, 100, 150, 250, read length is uniform (all reads have the same length)
-
Single and paired reads
-
Typical errors: generally high quality
-
Throughput: 540 Mb – 600 Gb
-
Runtime: 4 hours – 11 days
-
Cost: MiSeq – $125,000 (US list price in 2012)**
-
-
Summary: relatively short reads (but they can be paired), relatively slow, high throughput, high quality, high price.
-
Additional resources:
Ion Torrent
-
You can also take a look at the Ion Torrent subpage on the Life Technologies site here.
-
Technology videos:
-
Reads:
-
Read length: around 35 or 200 or 400 bps
-
Single reads (paired end reads have been introduced in 2012, but are still not commercially available).
-
Typical errors:
-
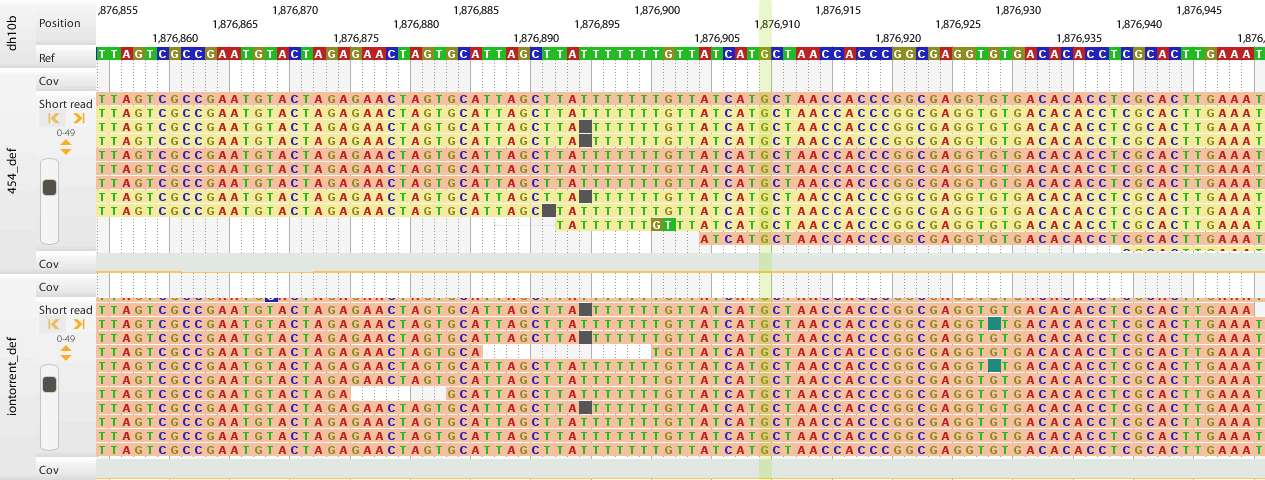
high amount of homoploymer errors
-
lower quality than 454 or Illumina
-
even lower quality at read ends
-
-
Throughput: from 3 Mb to 1.5 Gb (up to 10 Gb for Ion Proton), depending on read length and the chip used (for details see here).
-
Runtime: 0.5 – 7.3 hours
-
Cost: Ion Torrent PGM – $80,490 (US list price in 2012)**
-
-
Summary: long reads, high-throughput, fast, relatively cheap, but loads of homopolymer errors (over 1%) and lower quality in general.
-
Additional resources:
Roche 454
-
Technology videos:
-
Reads:
-
Read length: varies within a same run, the mean read length, depending on the instrument, is around 400-500 or 600-700 bps (theoretically read length can go up to ~1000 bp for GS FLX ).
-
Single and paired end reads
-
Typical errors:
-
homopolymer errors (lower percentage than in IonTorrent) (for details see here).
-
lower quality at read ends
-
-
Throughput: from 35 Mb (GS Junior) to 700 Mb (GS FLX Titanium XL )
-
Runtime: 10-23 hours
-
Cost: 454 GS Junior – $108,000 (US list price in 2012)**
-
-
Summary: longest reads, good quality, but low throughput and expensive, plus homopolymer errors are typical.
-
Additional resources:
Honorable mentions:
* Of course, a great amount of development, improvement and testing can be done by “in-company” teams, but in my opinion a substantial user base is crucial for proper, unbiased validation and testing of these sequencing tools.
** Prices were collected from the following article:
Liu et al., (2012) Comparison of Next-Generation Sequencing Systems Journal of Biomedicine and Biotechnology, Volume 2012, Article ID 251364, 11 pages, doi:10.1155/2012/251364, http://www.hindawi.com/journals/bmri/2012/251364/
Special thanks to Tibor Nagy for collecting the videos!