The UCSC Genome Browser is developed and maintained by the Genome Bioinformatics Group at the University of California Santa Cruz. The website contains a large number of reference sequences and genome assemblies from different species. Sequences and annotation sets can be downloaded or viewed in the Genome Browser.
A little more about the Genome Browser:
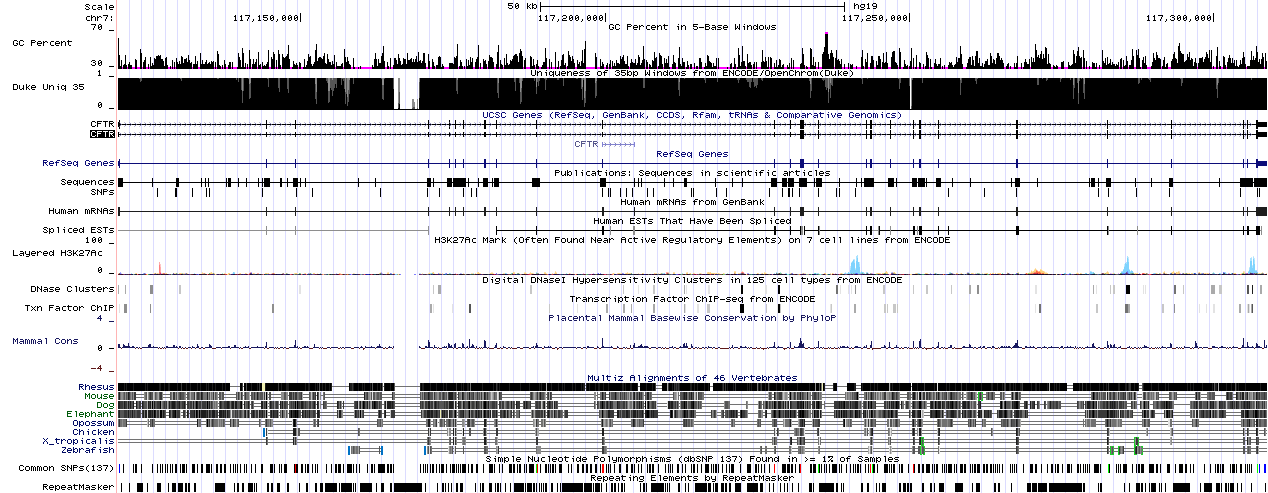
Additional to the reference sequence/assembly, there are numerous tracks available in the browser and you can also display your own track (if you are interested in adding a custom track, take a look at this page).
Let’s take a quick look at the human version of the browser!
You can find the VERY long list of available tracks for the latest human genome assembly (GRCh37/hg19) in the release log. A detailed help page is available for each track (just click on the name of the track). You can hide or show any of the available tracks and even the “density” of the visualisation can be selected.
Other than the default tracks (e.g. Refseq Genes), I usually use some version of the mapabilty track, usually the Duke Uniqueness track, which shows the uniqueness of a region in the genome. This information can be extremely useful for identifying alignment problems caused by similar/identical regions in the reference.
The other track I often use is GC percent, which shows the GC content of the sequence in five base long windows. Again, when you have unexpected low (or zero) coverage regions in an alignment, it’s usually wise to check out the GC-content of the reference sequence in the region. This is important, because moat sequencing technologies have trouble with very high or very low GC-content.