
Splicing is prevalent in MHC / HLA genes also, we just started to understand how the its machinery works, what are the steps in exon definition, and the role of enhancer and silencer motifs. Looking at the 3D structure of the yeast spliceosome we can see the difficulties structural chemists are facing: the RNA conformation is complex, unstable and difficult to crystallize. Nevertheless, it gives insights about the conformation changes you can have during splicing steps.
Also, to understand aromatic stacking and the role of non-Watson-Crick base pairs, it is good to practice your eyes in one of the largest RNA structure solved so far: the bacterial ribosome. In the pictures you can see some of us are really into the stacking of pactamycin (PDB structure 1HNX).
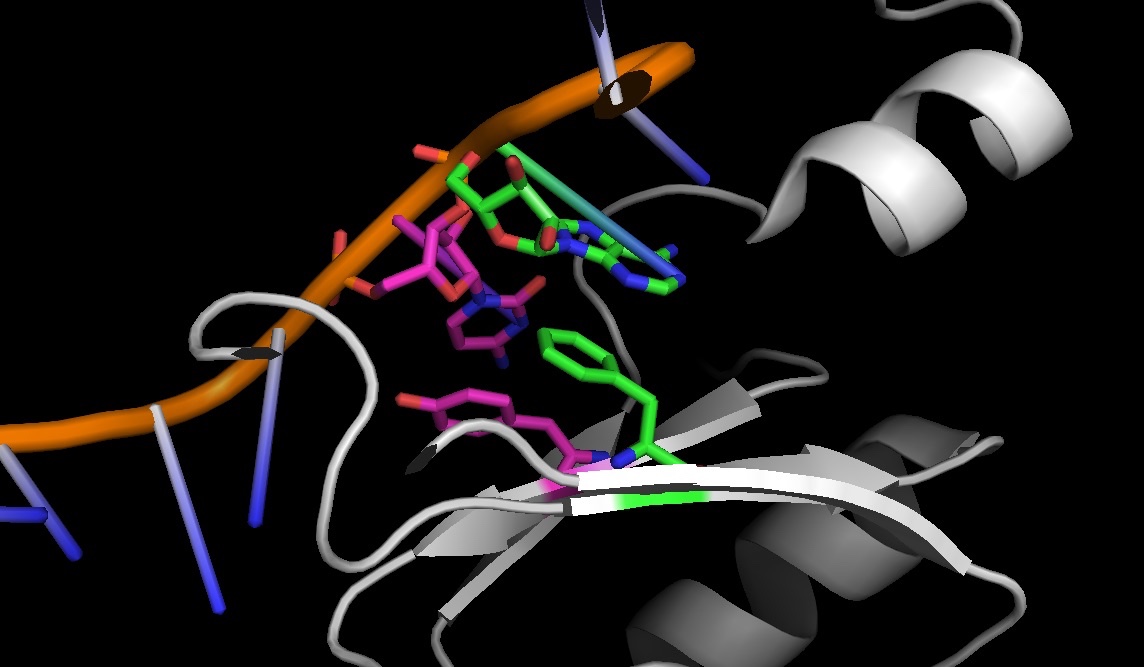
4PKD: stacking between amino acids and nucleic acids
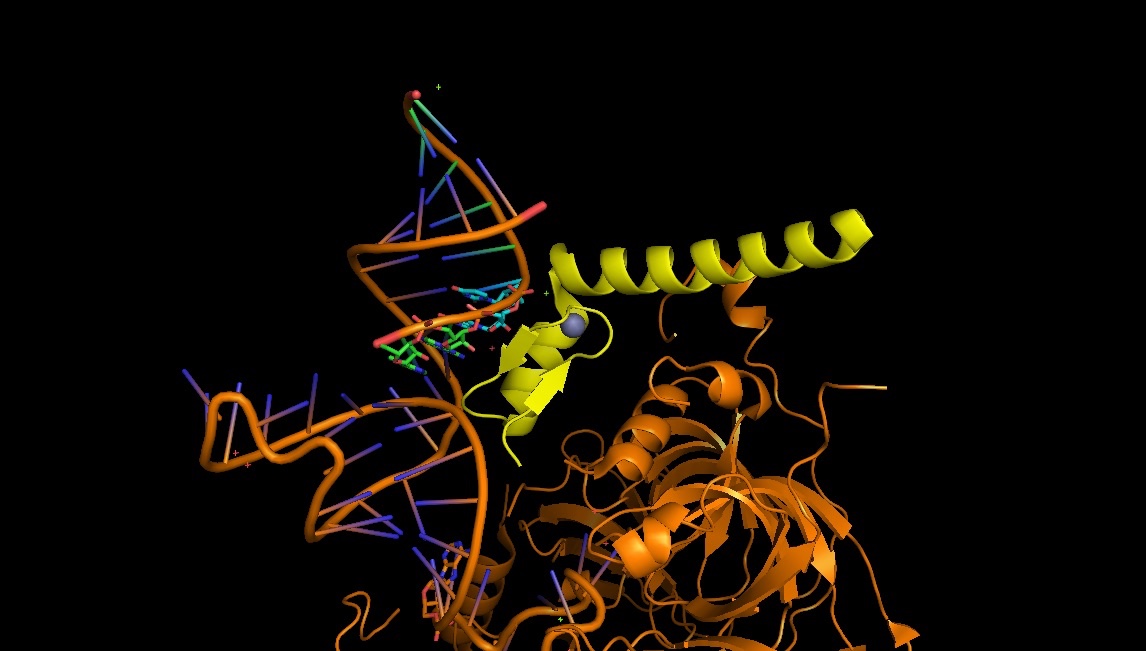
4PJO: the Zn-finger nucleic acid recognition part in U1 snRNP
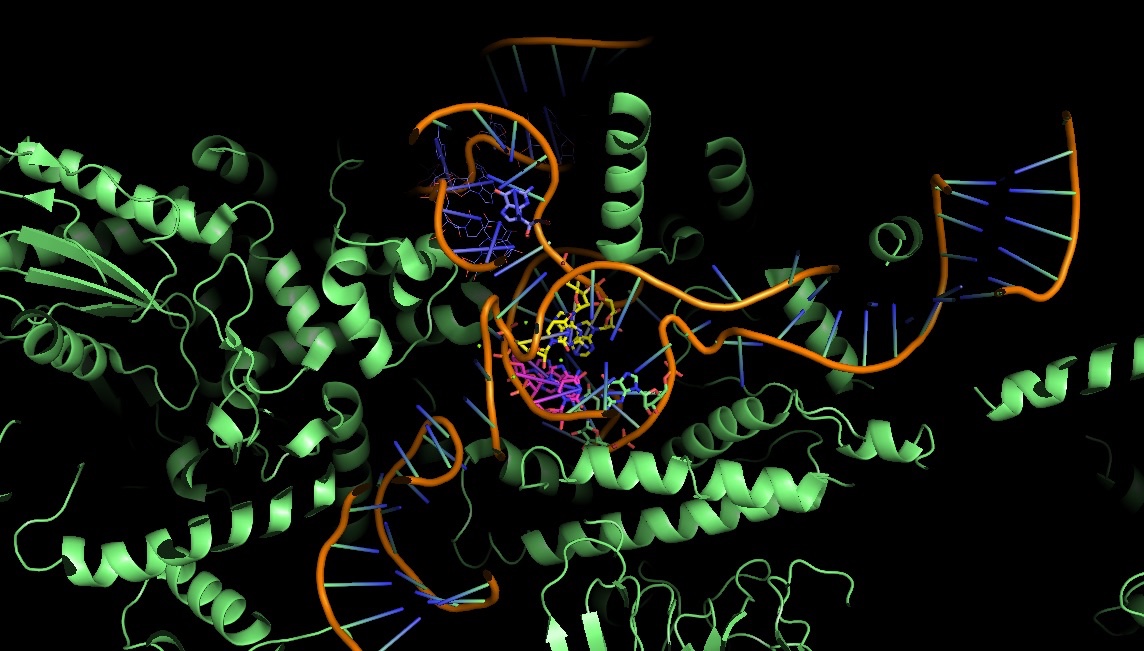
3JB9: the catalytic core of the yeast spliceosome: non-Watson-Crick base pairs are doing the job
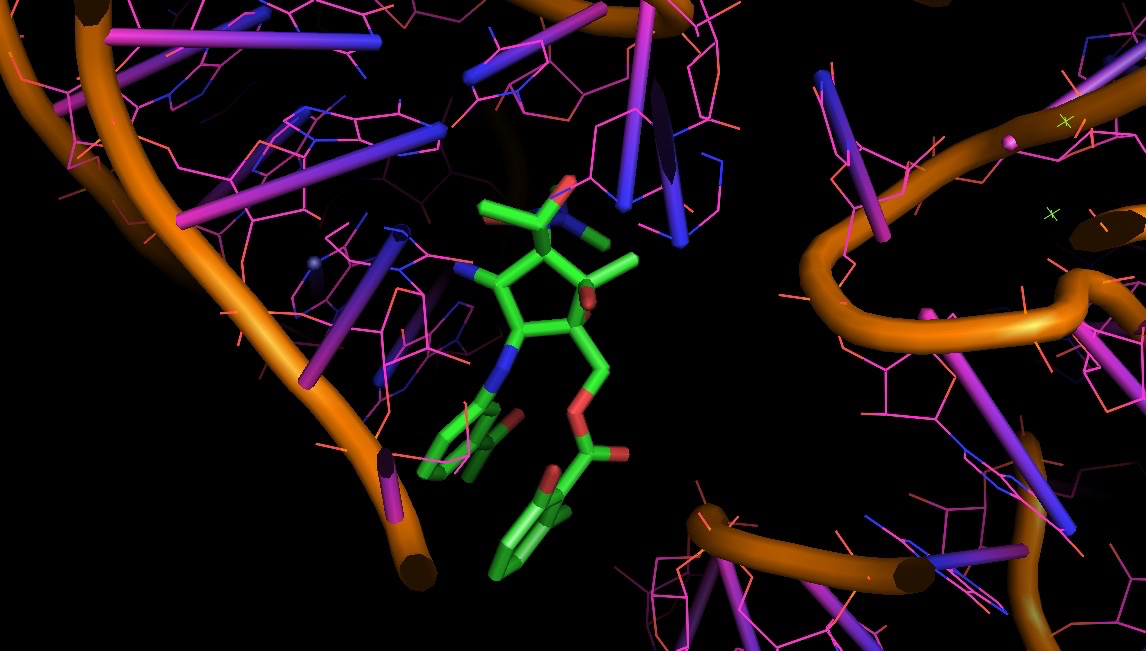
1HNX: pactamycin bound to bacterial ribosome: a test case to look at H-bonds and aromatic stacking
