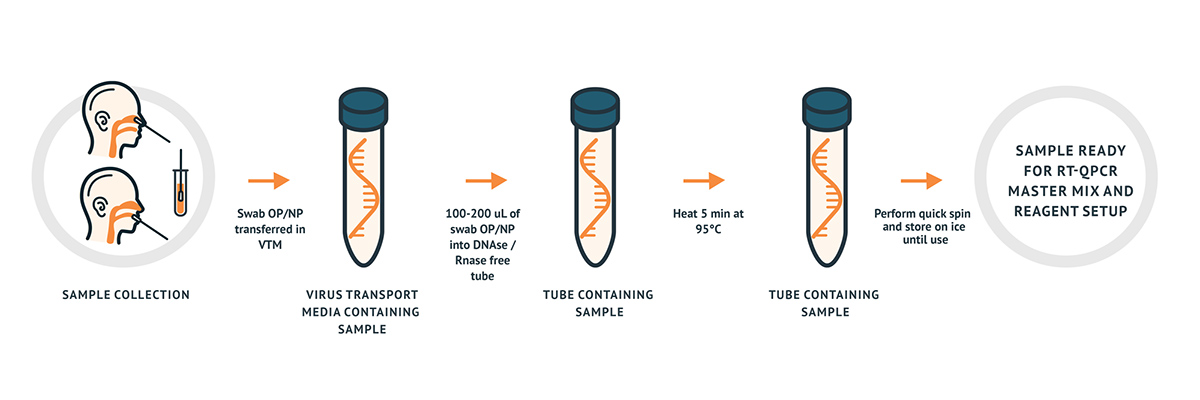
The AzureSeq is a CE, resp. RUO RT-qPCR test intended for the qualitative detection of nucleic acid from the 2019-nCoV in nasopharyngeal (NP) and oropharyngeal (OP) swabs from individuals with signs and symptoms of infection who are suspected of COVID-19. The assay can be used in combination either with an RNA extracted sample (indirect method) or directly with a sample collected into the viral transport medium (VTM).
Product Highlights
| Regulatory Status | CE-IVD |
| Extraction Principle | Direct Extraction |
| Target Genes | N1, N2 capsid and RNAseP |
| Turn Around Time From Sample to Result | Less than 120 min |
| Sensitivity | Low copy number (0.5 copies/μL) |
| Number of Pipetting Steps | 8 steps |
| Instrument Compatibility | BioRad CFX96, Roche Lightcycler 480 System and QuantStudio 3, 5, and 7 Pro |
Value Added
- Enable daily processing of hundreds of samples
- Accurate detection with reduced complexity, time and waste
- User-friendly, custom formats
- Increase your throughput 25% to 300%
- Fewer steps reduce potential user error
- Excellent for automation
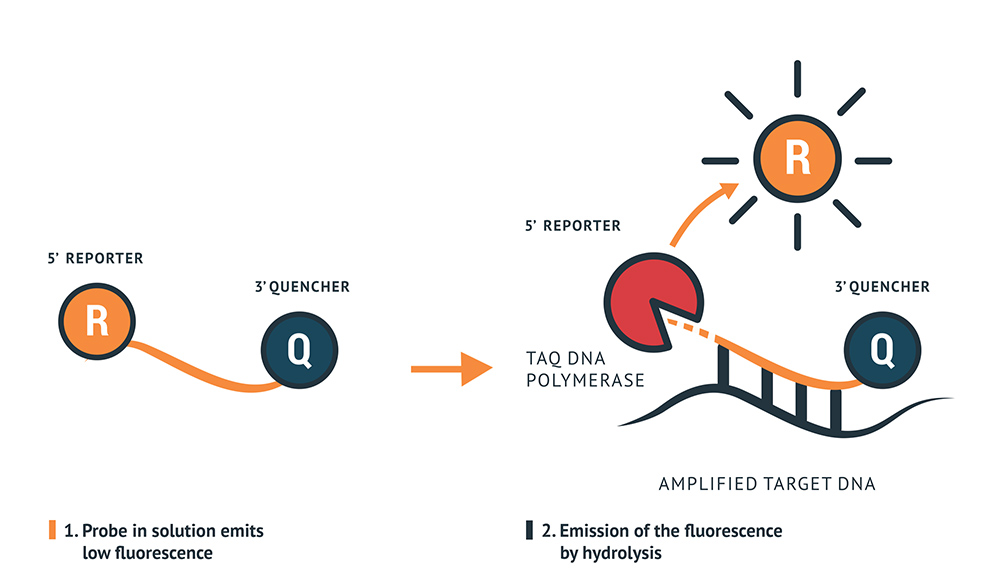
Detection Principle
The RT-qPCR Mastermix contains primers to amplify the specific genes regions of the virus and probes designed to anneal to a target sequence located between the forward and reverse primers. The probes contain a reporter dye at the 5’ end and a quencher dye at the 3’ end. When the reporter and quencher dye are in close vicinity, the fluorescence of the reporter dye is quenched by the quencher with very low fluorescence emission. During the extension phase of the PCR cycle, the polymerase will hydrolase the reporter dye, which will be released to emit a strong fluorescence. This fluorescence will increase by each cycle of polymerisation.
Kit Compatibility Format
| AzureSeq - 200 CE | 200 | In Solution: InhibiTAQ Plus HotStart Mastermix, RTScript, Target Specific Primers and Probes |