With “short” reads getting longer and longer, read trimming seems to be more and more useful. In my experience, long reads tend to have a significant quality drop at the read end, regardless of sequencing platform, so it makes sense to trim these low quality parts off.
Most tools that we used for read filtering last week can do read trimming as well.
First, let”s take a look at the per-base sequence quality of our example datasets!
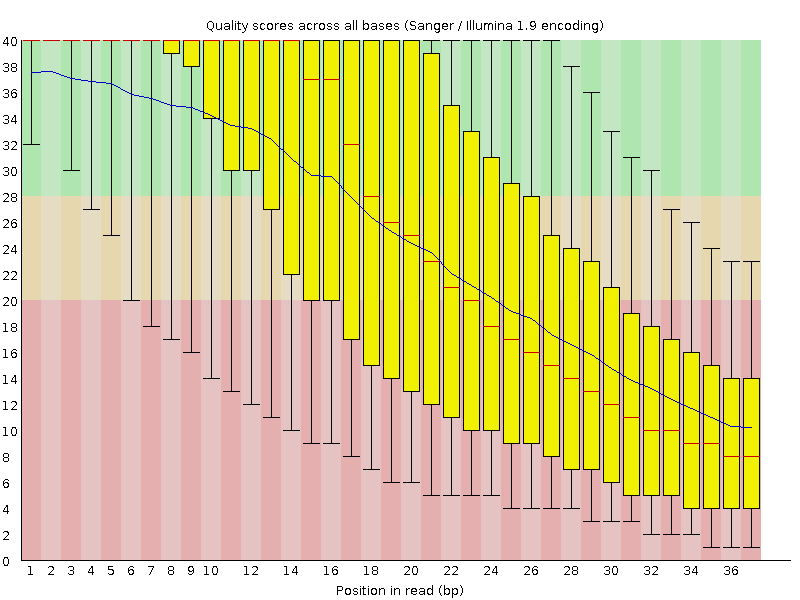
Per base quality diagram of the “historical” Illumina example data set produced by FastQC. The overall quality is quite low and there”s an almost exponential drop, but the reads are so short that there”s not really much to work with “trimming wise”. (Also, looking at this data, I”m still amazed about the progress the NGS industry made in the last few years).
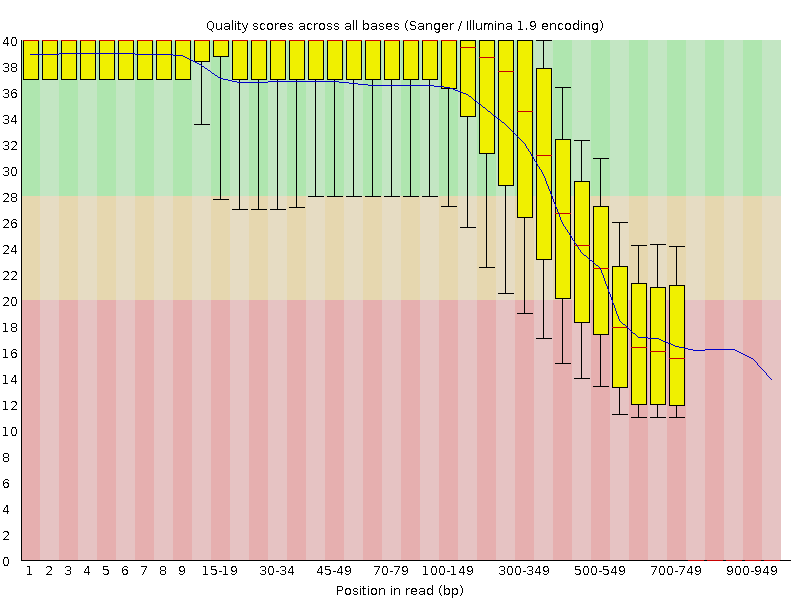
Per base sequence quality of the 454 example data produced by FastQC. Note, that the lower quality of the last bases is, at least partially, related to the lower number of very long reads.
The Roche data looks like a good candidate for read trimming. Let”s try a few different methods!
The easiest way to do read trimming is to remove a specific number of bases, regardless of the quality of these nucleotides. Let”s remove the last 50 nucleotides!
perl prinseq-lite.pl -fastq SRR797242.fastq -trim_right 50 -out_format 3 -out_good SRR797242_right_trim_50n Input and filter stats: Input sequences: 1,385,764 Input bases: 597,869,227 Input mean length: 431.44 Good sequences: 1,381,200 (99.67%) Good bases: 528,602,214 Good mean length: 382.71 Bad sequences: 4,564 (0.33%) Bad bases: 207,013 Bad mean length: 45.36 Sequences filtered by specified parameters: trim_right: 4564
You can also remove all bases after a specific number (basically trim all reads to a maximum length). As an exercise, let”s trim all read to a maximum of 800 bases:
fastx_trimmer -Q33 -l 800 -i SRR797242.fastq -o SRR797242_lenmax800.fastq
The best way for read trimming is most likely quality based trimming. Prinseq can do this as well:
perl prinseq-lite.pl -fastq SRR797242.fastq -trim_qual_right 15 -trim_qual_left 15 -out_good SRR797242_qual_trimmed_15 Input and filter stats: Input sequences: 1,385,764 Input bases: 597,869,227 Input mean length: 431.44 Good sequences: 1,385,764 (100.00%) Good bases: 597,019,714 Good mean length: 430.82 Bad sequences: 0 (0.00%) Sequences filtered by specified parameters: none






